Counting is vital in nature. Counting chromosomes is something that most animals, plants and even single-celled organisms need to know how to do to assure viability and to reproduce. Today, a team of geneticists reveals a remarkable mechanism that enables plants to count their chromosomes, solving a century-old mystery.
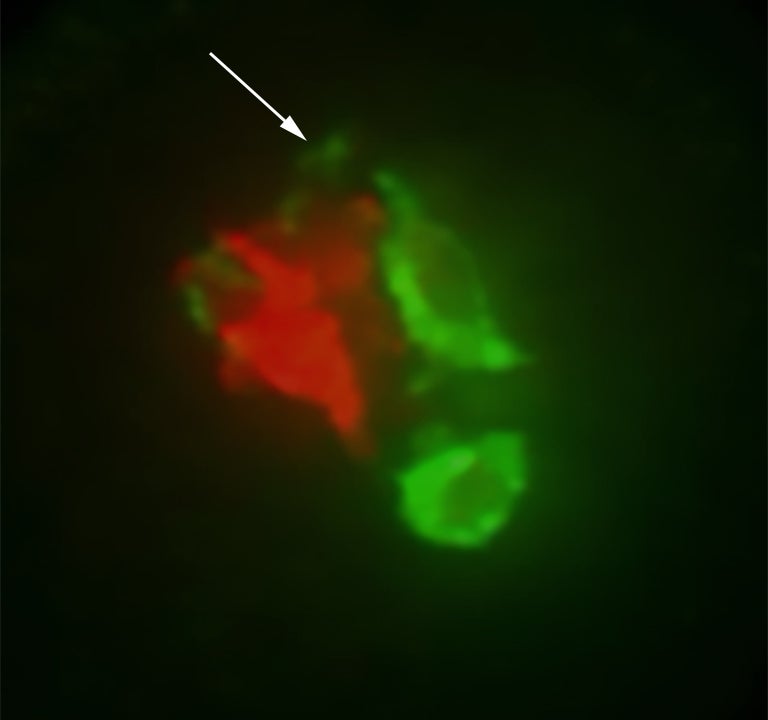
Inside a single pollen grain: a faint arc (arrow) suggests a connection between interior, or cytoplasm, of sperm (green) and the grain’s vegetative nucleus (red). It is thought that easiRNAs move from the vegetative nucleus into sperm via such connections.
Being able to count the amount of genetic material in a given cell or organism is a prime factor in ensuring that each cell receives the same complement of genes after each cell division. In plants, the mechanism that prevents individuals with imbalances in chromosome number from reproducing is something scientists call “the triploid block.”

The mechanism behind this classical problem now has been solved by Professor Rob Martienssen (pictured) of Cold Spring Harbor Laboratory (CSHL) and the Howard Hughes Medical Institute, and a team led by Filipe Borges, a postdoctoral researcher in his lab, with Dr. Claudia Köhler and colleagues at the University of Uppsala in Sweden.
Their research stems from the knowledge that chromosomes are covered with transposable elements (TEs), commonly called jumping genes. This raised the question, says Martienssen, “of whether a cell might be able to detect and count transposons as a proxy for chromosomes.”
One way of doing so, he reasoned, might have to do with small RNA molecules, which are generated by cells to keep potentially mutagenic transposons from becoming active.
Martienssen and Borges discovered a special kind of small RNA molecule found only in the pollen of flowering plants, called microRNA 845 (miR845). In a series of experiments reported in Nature Genetics the team now shows that miR845 seeks out and targets most transposons in plants genomes, triggering synthesis of secondary small RNAs called easiRNAs (epigenetically activated small interfering RNAs).

Arabidopsis seeds that abort because of the triploid block.
The more chromosomes a pollen grain has, the more transposons, and therefore the more easiRNAs it accumulates in its sperm. When the sperm unites with the female gametes (eggs) in a developing seed, a kind of counting ensues. If the “dosage” of chromosomes and easiRNAs is far out of balance, it triggers the triploid block and the seed collapses.
The team proposes a mechanism behind this block, as easiRNAs are able to silence key genes called imprinted genes on both the maternal and paternal sides. Such silencing results in the sterilization of the emerging seed. The discovery adds to basic biological understanding of chromosome regulation. It could also be a boon to plant breeders, who, explains Martienssen, would like to figure out ways to get around the triploid block, to cross otherwise reproductively incompatible plants.
Funding
National Institutes of Health; National Science Foundation Plant Genome Research Program; Howard Hughes Medical Institute and Gordon and Betty Moore Foundation; CSHL Cancer Center Support Grant; European Research Council Starting Independent Researcher grant; Swedish Science Foundation; Knut and Alice Wallenberg Foundation; Marie Curie IOF Postdoctoral Fellowship.
Citation
Borges, F et al, “Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis” appeared online in Nature Genetics January 15, 2018.
About Cold Spring Harbor Laboratory
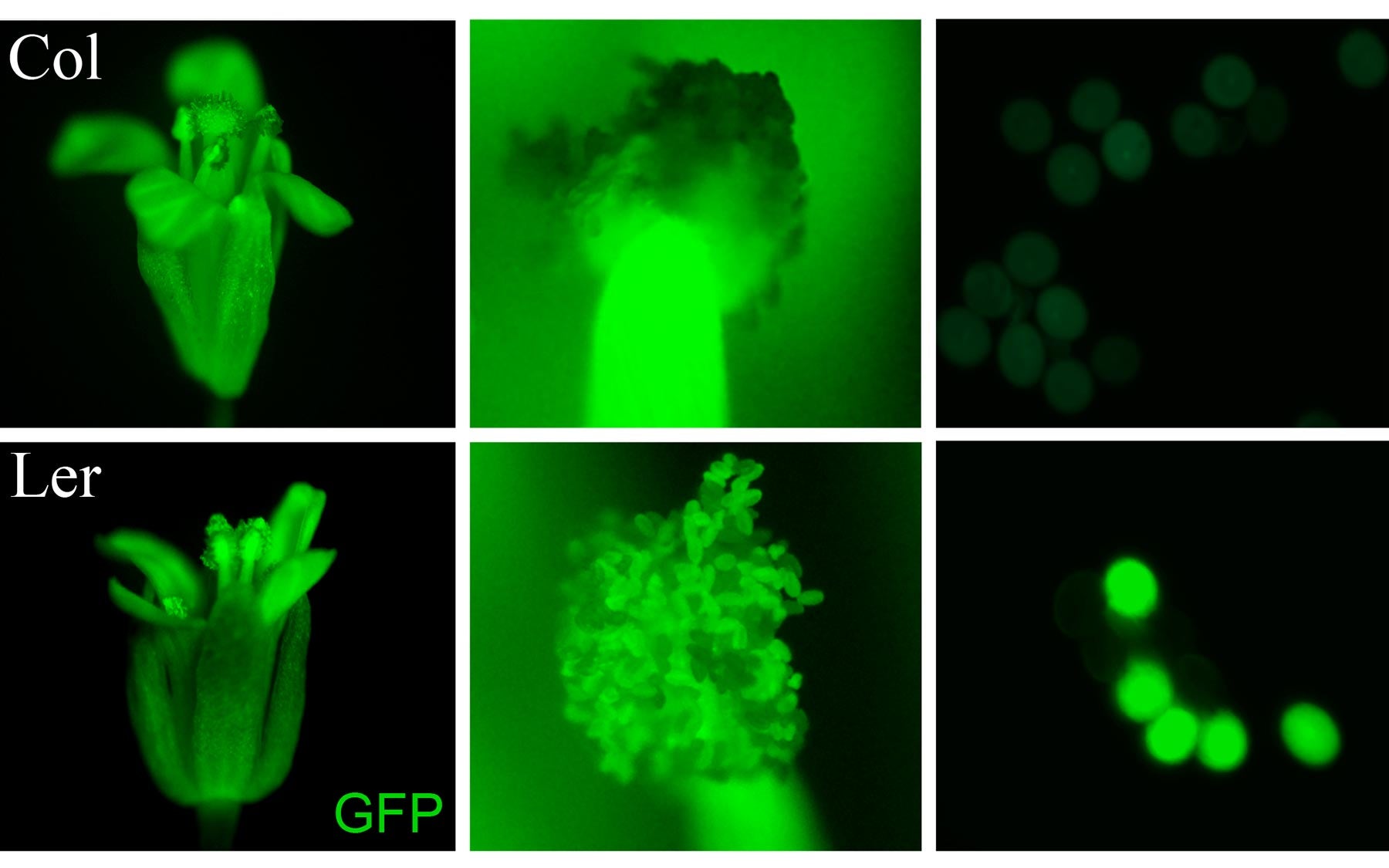 miR845 is expressed in pollen grains, a key part of the mechanism that enables flowering plants to count their chromosomes. The two rows show variants of wild-type Arabidopsis plants. In the Arabidopsis variant top row, miR845 is expressed, and pollen grains (top right) are invisible. In the variant in the lower row miR845 is absent, and pollen grains are visible (lower right). The triploid block will only affect the plant in the top row.
miR845 is expressed in pollen grains, a key part of the mechanism that enables flowering plants to count their chromosomes. The two rows show variants of wild-type Arabidopsis plants. In the Arabidopsis variant top row, miR845 is expressed, and pollen grains (top right) are invisible. In the variant in the lower row miR845 is absent, and pollen grains are visible (lower right). The triploid block will only affect the plant in the top row.










