Berkeley, California, USA
February 4, 2019
From mystery to discovery: CasX becomes “Cas!!!”
Scientists use the letter “X” to designate the mysterious (such as X-rays, and the X chromosome, when both were first discovered). Meet the newest entry, CasX, the mystery of which was cracked by a team of “detectives” at the Innovative Genomics Institute. Two landmark discoveries of the past century – recombinant DNA, and gene editing with CRISPR-Cas proteins– came out of the study of microbes, which is where CasX, an initially puzzling member of the CRISPR family, was first found. CRISPR-Cas is like a word processor for DNA of living cells, and gives biomedical scientists, agronomists, and clinicians an “all access” pass to engineering cells and organisms. In all technologies, the excellent is a healthy rival of the good. Published today in Nature, scientists have discovered that CasX, which is physically smaller than other editors, and has a different shape, punches well above its weight and is a formidable gene editor. Size does matter, and small is better – if genetic engineering is the task at hand.
Teaching a new dog tricks both old and new
In word processing, we use a mouse to click on the spot we want to edit. The enzyme CRISPR-Cas9 is found in near every biology lab in the world because it can be directed to “click” on a gene of interest using an astonishingly simple principle: give it a small piece of RNA that matches the gene, and the enzyme will find that gene in a living cell and cut it. This allows scientists to swap, insert, or delete DNA sequences with remarkable ease. In 2012, Cas9 became the first such enzyme to be discovered and adapted for this purpose. Since then, scientists have discovered multiple other Cas (CRISPR-associated) proteins. Just as the Berkeley cyclotron led to the discovery of new chemical elements, many new Cas proteins were found at UC Berkeley as well. In December 2017, the laboratories of Jennifer Doudna and Jill Banfield discovered two new types of CRISPR proteins that were quite unlike all the previous ones; they were named CasX and CasY, since it was not clear what they actually did. Could these mysterious proteins possibly compare to, or rival, their famous predecessor, Cas9?.
Is the excellent not an enemy of the good? Not in technology development. “While the current proteins like Cas9 and Cas12 are robust in a research setting, what is important in the clinic is actually quite distinct from the lab,” says co-lead author Ben Oakes, who runs a research group through the IGI’s Entrepreneurial Fellows Program, and focuses on developing practical applications of CRISPR. Oakes was optimistic that CasX could be a solution to a known challenge: other CRISPR-based editors are physically large and come from bacteria that infect human beings. For use in the clinic, this means that scientists need to face logistical hurdles of delivery and immunogenicity.
Viruses are used as medical “Trojan horses,” sneaking therapeutic DNA into diseased cells.
Scientists and physicians who work on gene editing as a disease therapeutic have a somber joke: what three things matter in gene editing? Delivery, delivery, and delivery. If you cannot place the gene editor inside your target cell or organ, you will fail. To introduce Cas9 into specific cells in the body, scientists often use a molecular delivery truck: a carrier virus called AAV. While nimble, AAV has limited carrying capacity, and this means having to deal with tightly packaging the necessary machinery or instructions for genome editing. CasX is quite a bit (~40%!) smaller than the most commonly used gene editor, Cas9, and this means that packaging CasX into AAV vessels could leave room for other tools and genome editing accessories. What if the small size of CasX comes with a penalty on what it can do? Scientists now definitively show that CasX is a potent and efficient gene editor and gene regulator; thus, the gene editing toolbox now has a powerful fighter not only in its featherweight class but in all categories.
The small size of CasX is not its only noteworthy feature: so are its origins – CasX was found in a bacterium that human beings are never exposed to. Why is this interesting? Many of the best Cas9 proteins used for genome editing come from bacteria that commonly reside on or inside human bodies; as a result, a number of people carry an immune memory of being exposed to Cas9. What will happen in a human being with such a memory who is treated with Cas9 for gene editing? The answer is not known but is on the mind of all who are using Cas9 for clinical trials, a number of which are on their way.
“The immunogenicity, delivery, and specificity of a genome-editing tool are all vitally important,” says Oakes. “We’re excited about CasX on all of these fronts.”
In a league of its own
After determining that CasX can edit DNA, the research team set out to understand how this mighty machine works and how they might be able to improve it. Similar to renovating a house, an engineer must have a detailed blueprint before rearranging any pipes or cables, so the team set off to map out such a blueprint.
At first glance, CasX appears to act like its predecessors Cas9 and Cas12; the fine-resolution blueprint of the enzyme tells a different story. Co-lead authors Jun-Jie Liu and Natalia Orlova used a tour-de-force approach, known as cryo-electron microscopy: they took hundreds of thousands of images of CasX and combined them to build an atom-by-atom portrait of it. The result was not a mere still-life: Liu and Orlova captured snapshots of the CasX protein going through the motions of editing a gene. This atomic movie gives essential clues for how to use CasX in editing.
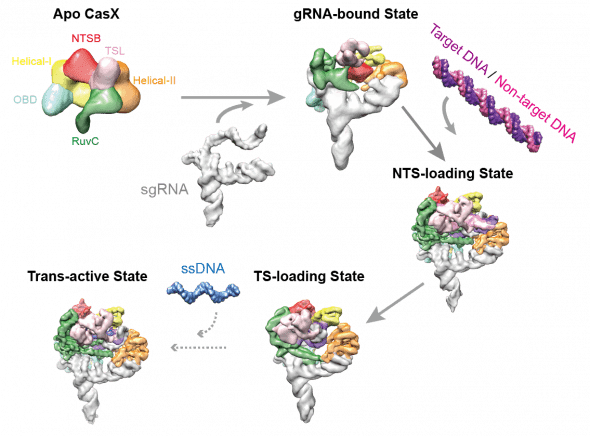
Electron microscopy snapshots of CasX in action.
Assembling film revealed a stunning feat of evolution. A dolphin looks like a fish – but it is a mammal, ancestors of which evolved“fishiness” quite independently of actual fishes. By the same token, CasX precisely cuts both strands of the DNA double helix, just like Cas9 and related enzymes; but this a skill it appears to have learned on its own. Based on the protein’s unique molecular makeup and shape, CasX evolved independently of Cas9, sharing no common ancestor. “The first thing that jumps out is how the highly unique domains accomplish similar roles to what we have seen with other RNA-guided DNA-binding proteins. CasX’s minimal size, with no fat on the bone, helps to clearly demonstrate there is a basic recipe that nature uses,” says Oakes. “Understanding this recipe will help us to better evolve and engineer genome editing tools for our purposes rather than nature’s.”
Expanding and exploiting CasX
Beyond settling for what nature has evolved, Oakes plans to engineer CasX for specialized functionality and improved editing efficiency. “We’re very excited to build on nature’s design and engineer new, more effective CRISPR proteins, as our recent work on ProCas9 has demonstrated. Developing CRISPR proteins for specific purposes will vastly expand the reach and value of CRISPR technology to society.” The story continues as Oakes and his colleagues begin to test out other homologs, or flavors, of CasX proteins, identifying promising candidates for therapeutic applications.
Collaboration amongst a diverse set of research groups at the IGI and across the UC Berkeley campus has led to an exciting pipeline of CRISPR protein discovery and repurposing into genome editing tools. “The culmination of biochemistry, genome editing, and structure experiments within this single study is a prime example of the comprehensive efforts that are underway at the IGI,” says Jennifer Doudna, IGI’s Executive Director. “We aren’t just looking to uncover the next pair of molecular scissors. We want to build the next Swiss army knife.”
–
This study was published in the online edition of the journal Nature as “CasX enzymes comprise a distinct family of RNA-guided genome editors.” In addition to Oakes, Liu, and Orlova, other authors are Enbo Ma, Hannah B. Spinner, Katherine L.M. Baney, Jonathan Chuck, Dan Tan, Gavin J. Knott, Lucas B. Harrington, Basem Al-Shayeb, Alexander Wagner, Brett T. Staahl, Kian L. Talyor, Julian Brotzman, Eva Nogales, and Jennifer Doudna. This research was funded by the Innovative Genomics Institute’s Entrepreneurial Fellows Program, the National Institutes of Health, the Howard Hughes Medical Institute, and the National Science Foundation.
The Innovative Genomics Institute (IGI) is a non-profit, academic partnership between UC Berkeley and UC San Francisco that supports collaborative research projects across the Bay Area. The IGI’s mission is to develop and deploy genome engineering to cure disease, ensure food security, and sustain the environment for current and future generations. As pioneers in genome editing, functional genomics, and other cutting-edge technologies, IGI scientists continuously push the boundaries of science.