Bloomington, Indiana, USA
February 16, 2022

Roger Innes. Courtesy photo
Plants can recognize pathogens and respond to them. Roger Innes, Distinguished Professor in the Indiana University College of Arts and Sciences Department of Biology, and members of his lab are investigating the molecular and cellular basis of disease resistance in plants. They want to understand how plants detect pathogens and how detection then activates immune responses.
Innes lab researchers and colleagues in the lab of Blake Meyers, a member of the Donald Danforth Plant Science Center in St. Louis and Professor of Plant Sciences at University of Missouri-Columbia, recently discovered that the leaves of plants accumulate RNA on their surfaces and in the spaces between cells.
RNA is usually thought of as a molecule that can direct cells to synthesize specific proteins. An example that has frequented the news in the last two years is the messenger RNA (mRNA) in COVID-19 vaccines that directs human cells to make SARS-CoV2 spike protein. Some RNAs, however, serve other functions.
To prevent infection by disease-causing microbes, plants secrete diverse antimicrobial compounds into their extracellular spaces. Included among these compounds are small RNA molecules 21 to 24 nucleotides in length (sRNAs) that can be taken up by microscopic organisms such as bacteria. The sRNAs are thought to cause the destruction of mRNAs with complementary sequences. The processes by which sRNAs are secreted, how they are protected from degradation, and how they are taken up by pathogenic microbes are all poorly understood.
Although extracellular sRNAs had been shown to co-purify with extracellular vesicles (EVs), it had not been clearly established whether sRNAs were located inside or outside EVs, or whether plants secreted RNAs longer than 24 nucleotides.
Researchers in the Innes lab isolated EVs from the extracellular spaces in the leaves of Arabidopsis thaliana (a species of mustard plant) and then treated the preparations with enzymes that degrade proteins and then with different enzymes that degrade RNA. The RNA remaining after these treatments was then analyzed by RNA sequencing performed by the IU Center for Genomics and Bioinformatics and the Meyers laboratory. These analyses revealed that sRNAs are associated with protein complexes located outside EVs. Significantly, they found that Arabidopsis secretes both sRNAs and much longer RNAs, from 30 to over 500 nucleotides in length. The longer RNAs do not code for proteins, and many have a circular structure. Notably, both the long non-coding RNA and the small RNAs were found to be highly enriched in a post-transcriptional modification known as N6methyladenine (m6A). The researchers speculate that this modification might be required for secretion of RNA.
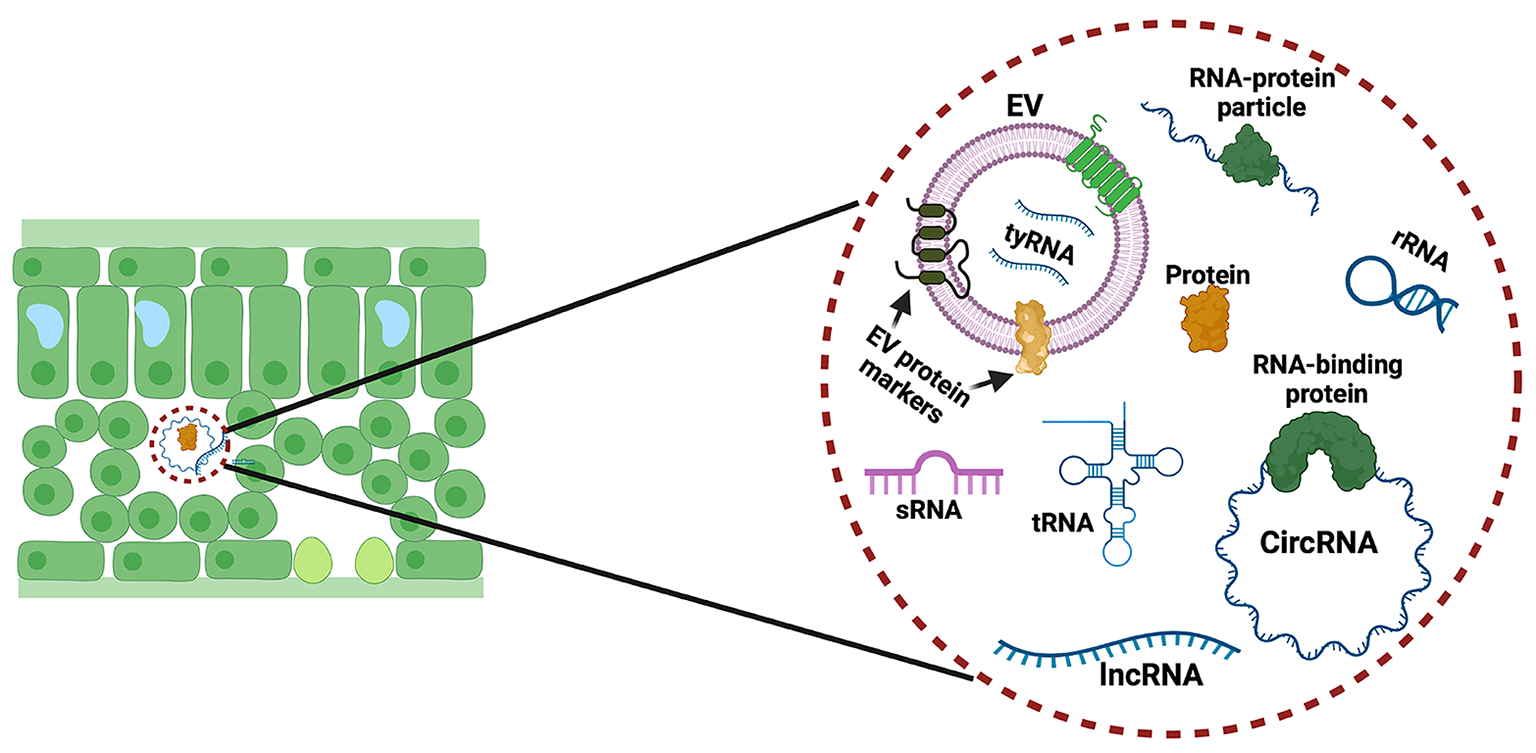
Plant cells secrete diverse RNA species—including small RNAs, long non-coding RNAs, and circular RNAs. Illustration courtesy of the Innes lab
A paper discussing the findings has just been published online in the scientific journal The Plant Cell.

Hana Zand Karimi. Courtesy photo
First author on the paper is Hana Zand Karimi, Ph.D. 2021, who was a graduate student in the Innes lab at the time of the study. The journal includes an "In Brief" article highlighting the paper and incorporating a model figure provided by Zand Karimi. Other authors on the paper are Brian Rutter, Lucía Borniego, Kamil Zajt, and Innes from IU as well as Patricia Baldrich and Meyers from the Donald Danforth Plant Science Center.
That plants secrete long non-coding RNAs, including circular RNAs, was an unexpected discovery and raises the question as to why. Do they play a role in cell-to-cell communication within the plant? Are they an important component of the immune system? The researchers hope to understand how these RNAs are secreted and the roles RNA-binding proteins and post-transcriptional modifications play in the process.
The National Science Foundation has granted Innes and his Meyers lab colleagues $2.3 million to investigate the role of extracellular RNA (exRNA) in the immune system of plants. They will test the hypothesis that exRNA functions to protect plants from infection by fungi and bacteria.
Innes and his colleagues have selected Arabidopsis, soybean, tomato, lettuce, pineapple, rice, and maize for the study. The species were chosen based on their phylogenetic diversity, genomic resources, importance as crops, and diversity in physiology. The researchers hope to learn whether specific exRNAs are broadly conserved across plant species as well as how exRNAs are secreted and whether post-transcriptional modifications are central to the process. They will seek to find out why plants produce exRNAs and whether they play a fundamental role in plant-microbe interactions.

Hana Zand Karimi injects soybean leaves with a bacterial pathogen to induce immune responses. Photo courtesy of the Innes lab
If Innes and his colleagues' investigation proves their hypothesis correct, it could provide them with a means to create crop plants with improved immune systems more resistant to disease. Such crops could feed the global population in a more sustainable manner.
The award was co-funded by the Plant Genome Research Program and the Plant Biotic Interactions Program in the NSF Division of Integrative Organismal Systems.
Innes's investigation into how extracellular RNA in plants can recognize and respond to pathogens has been featured in IU Research in the News. Listen to the podcast (starting at 1:58) or view the transcript (second story) in "Research Impact Episode 233" (February 23, 2022).
Find the podcast
View the transcript