Itahca, New York, USA
June 30, 2017
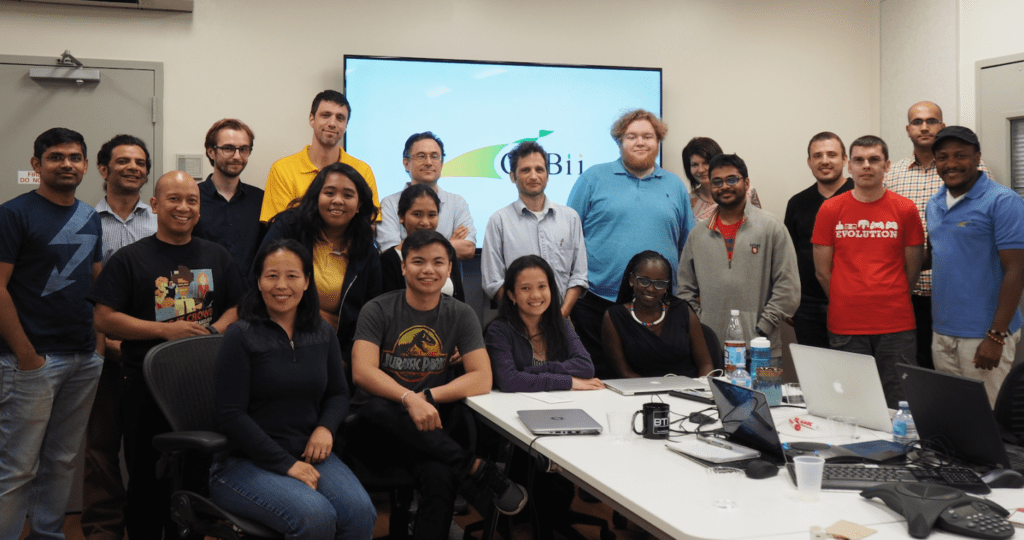 Participants in GOBII’s 2017 hackathon served as representatives from a diverse group of organizations, including Cornell University, Diversity Arrays Technology (DArT), CIMMYT, ICRISAT, IITA, IRRI, and the James Hutton Institute.
Participants in GOBII’s 2017 hackathon served as representatives from a diverse group of organizations, including Cornell University, Diversity Arrays Technology (DArT), CIMMYT, ICRISAT, IITA, IRRI, and the James Hutton Institute.
The GOBII project’s annual hackathon united 25 software developers, bioinformaticians, computational biologists, and application specialists on a week-long mission: not only were they focused on developing many improvements to their genomic data management system and key breeding decision support tools, but also on gaining as much face-to-face time with each other as possible.
GOBII, which stands for Genomic Open-source Breeding Informatics Initiative, seeks to develop a publicly accessible genomics database to enable public sector breeders to accelerate genetic gains initially to support five major staple crops: rice, maize, chickpea, sorghum, and wheat. GOBII’s annual hackathon plays a critical role in the development of high priority tasks while leveraging skills and capabilities across different collaborators throughout the week.
Not only did this year’s hackathon attendees serve as representatives of a diverse group of partner organizations, but also united diverse cultures from around the world including Kenya, Mexico, Australia, India, the Philippines, and the United Kingdom. Because of GOBII’s global reach, partners frequently work together online in smaller and more selective networks.
By hosting an annual hackathon at BTI, GOBII partners have a greater ability to improve their productivity versus an online-only working environment. “We work together frequently, but we often don’t have the connection channels that we do here,” according to Yaw Nti-Addae, GOBII’s software development group leader. “The most important aspect of this hackathon is that we get to work together face-to-face.”
The hackathon featured a mix of seasoned attendees and first-time participants, allowing contributors to learn from and teach one another while improving everyone’s individual abilities. “This [hackathon] gives us a better way to make improvements and create better systems together,” according to Andrew Kowalczyk, a first-time attendee and bioinformatician at Diversity Arrays Technology (DArT). “Although the web enables us to do our jobs remotely, interaction can still be limited. Being in-person provides greater transparency to our work, often exposing one group’s problem which another has already solved”.
GOBII’s ultimate goal is to develop genomic software tools that can help breeders access extensive genomic data to breed crops that are more disease resistant, drought tolerant, and higher yielding. Milcah Kigoni, a GOBII graduate intern and computational biology researcher at International Institute of Tropical Agriculture (IITA), believes the hackathon helps to keep project collaborators motivated.
“In Africa, we are working with resource-poor, small-scale farmers who rely heavily on crop farming as a source of food and income. If these farmers do not have to deal with crop diseases and other challenges such as drought-susceptible crops, they can have higher crop production, improved nutrition, increased income and subsequently enjoy better livelihoods. This has motivated me because I know that through genomics-assisted breeding, we can achieve this.”
This year’s hackathon focused on developing a second version of GOBII’s Genomic Data Management System, including associated technologies such as a quality control pipeline, Marker Assisted Back-Crossing (MABC), and Galaxy modules for genomic selection. Throughout the week, participants worked collaboratively to develop system improvements, updates, and develop additional functionalities and features to be included in the second version.
GOBII 2017 Hackathon

2017 Attendees and Organizations (June 19-24, 2017)
| Angel Villahoz–Baleta |
GOBII |
| Elizabeth Jones |
GOBII |
| Josh Lamos-Sweeney |
GOBII |
| Kelly Robbins |
GOBII |
| Kevin Palis |
GOBII |
| Monica Franciscus |
GOBII |
| Phil Glasser |
GOBII |
| Syed Raza |
GOBII |
| Yanxin Star Gao |
GOBII |
| Yaw Nti-Addae |
GOBII |
| Ramu Punna |
Cornell University |
| Andrew Kowalcyzk |
Diversity Arrays Technology (DArT) |
| Kate Dreher |
International Maize and Wheat Improvement Center (CIMMYT) |
| Victor Ulat |
International Maize and Wheat Improvement Center (CIMMYT) |
| Chaitanya Sarma |
International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) |
| Prasad Bajaj |
International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) |
| Selvanayagam Sivasubramani |
International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) |
| Milcah Kigoni |
International Institute of Tropical Agriculture (IITA) |
| Angel Manica Raquel |
International Rice Research Institute (IRRI) |
| John Carlos Ignacio |
International Rice Research Institute (IRRI) |
| Venice Juanillas |
International Rice Research Institute (IRRI) |
| Viana Calaminos |
International Rice Research Institute (IRRI) |
| Gordon Stephen |
James Hutton Institute |
| Iain Milne |
James Hutton Institute |