Berkeley, California, USA
April 17, 2023
Jennifer Doudna, co-developer of CRISPR genome editing, and Jill Banfield, pioneer of genome-resolved metagenomics, are heading the new Berkeley Initiative for Optimized Microbiome Editing (BIOME) at the IGI. But what exactly is a microbiome? What does it mean to edit one, and why would you want to? To help explain why the institute is so excited by this new line of research, we wanted to take a step back and introduce readers to these concepts, with help from Ben Rubin and Brady Cress, two IGI BIOME PIs.
For more in this series, check out Metagenomics 101 with another BIOME PI, Spencer Diamond.
What is a microbiome? Where are they found?
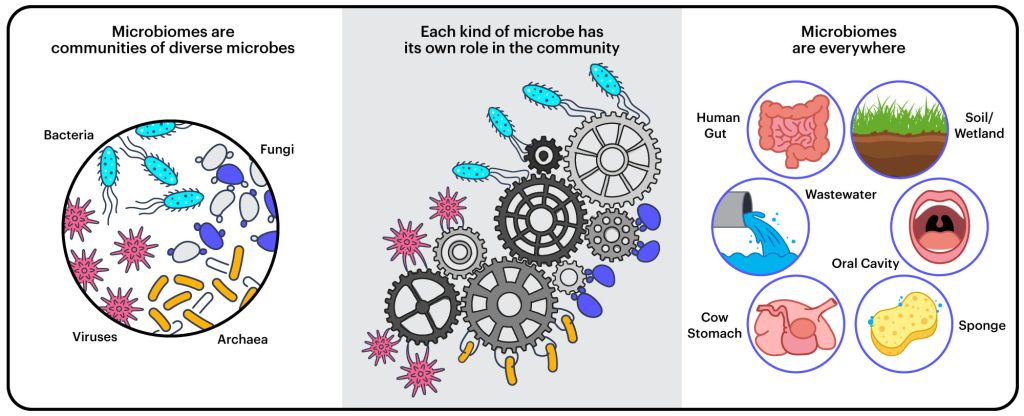
A microbiome is a community of microorganisms — bacteria, archaea, unicellular fungi, viruses, and other microbes — living and working together in a particular environment.
“It’s not just that they’re in the same place,” explains Rubin. “But their properties as a whole are greater than the sum of the parts.”
Microbiomes are found in all kinds of places — in water, soil, on and in and animals and plants — and they often provide important services to the ecosystem or the host organism.
How are microbiomes important for our health?
“One of the examples we think about a lot is the collection of microorganisms in your gut,” says Rubin. “They have a tremendous impact on everything from your weight to your neurological development.”
All together, there are at least as many microbial cells in the human body as human cells, and they have an impact on our body, some positive, some negative. To date, research correlates the human gut microbiome composition with cardiovascular disease, obesity, asthma, allergies, and neurological disorders like Alzheimer’s disease. The microbiome of the mouth influences not only oral health, but cardiac health, and the skin microbiome differences are linked to infection and inflammation.
“The key point is these microbes are interacting extensively with each other, and they’re also interacting with many human tissues and human cells. To date, a lot of studies on microbiomes have been really focused on correlations of diseases with the composition of a microbiome. In other words, investigating which microbes are present and how that impacts health,” says Cress. “The field has made a lot of hypotheses, but so far there aren’t a lot of really great genetic tools for testing these hypotheses. So we know that the microbiome is bound to be impacting our health to a very large degree, but we need to develop more ways to understand and affect it.”
 Brady Cress (left) and Ben Rubin (right)
Brady Cress (left) and Ben Rubin (right)
One of the first goals of BIOME is better understanding the role of the gut microbiome in childhood asthma in collaboration with Sue Lynch, the Director of the Benioff Center for Microbiome Medicine at UCSF. Childhood asthma affects 300 million children worldwide, particularly in low- and middle-income countries, and Lynch’s research has found a strong link between specific bacteria in the gut microbiome and the development of asthma.
Where else do microbiomes play a big role?
Soil microbiomes are a key focus of IGI research because of the connection with both storing carbon in the soil and the release of potent greenhouse gases, like methane. BIOME is working on developing genetic tools to edit the genomes of soil microbes in order to increase biological carbon sequestration and reduce methane release as part of IGI’s research on net-zero farming.
Soil microbes aren’t the only ones that release methane: microbes in livestock guts are the single largest source of human-generated methane emissions. In fact, the majority of human-made methane emissions originate from microbiomes, including in rice paddies, waste lagoon, and landfills.
Microbiomes also play roles in bioremediation and bioproduction.
“One very important microbial community is in water treatment,” says Rubin. “Both for processing sewage and treating drinking water.” Other examples of microbial bioremediation include breaking down oil in oil spills and reducing heavy metal contamination in soils.
“In terms of bioproduction, we’re working with the Joint Bioenergy Institute on producing fuel and other valuable products from biomass using microbial communities,” says Rubin. “The food industry also uses microbiomes. Every cultured food that you think about — yogurt, cheese, kimchi — has a microbiome role.”
What tools do we have right now for changing a microbiome?
Right now, our main tool for modifying microbiomes is antibiotics. While they are a critical and life-saving class of drugs, they’re “a blunt tool,” says Cress. Antibiotics wipe out whole populations of bacteria.
“It’s like taking a sledgehammer to the microbiome,” says Rubin. “But health is usually more complicated than that. It’s usually not just presence that is bad or absence that is good, but that’s how antibiotics affect a microbiome.”
There has also been considerable interest in probiotics to try to influence microbiomes. Probiotics are typically supplements that contain “good bacteria” used with the goal of promoting our health. Oftentimes, probiotics have little impact as they aren’t integrated into the microbial community, but are more like visitors just passing through.
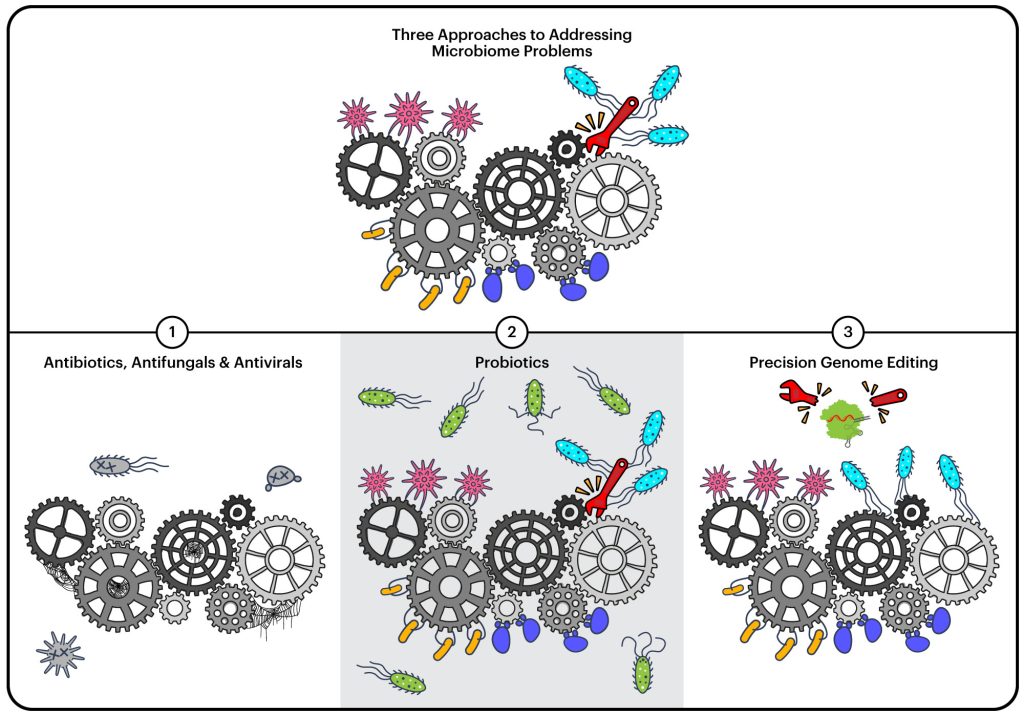
Fecal transplants have been used with success for treating challenging antibiotic-resistant infections but may not be applicable in many other indications. “This is actually where the stool from one human is used to replace and repopulate the microbiome of another human who’s suffering from a gut associated disease,” explains Cress. “Much like with probiotics, there’s limited efficacy and many of the same problems – it’s not clear how to make sure that the microbes that are transferred from one human to another actually persist and confer the same beneficial effects on gut health.” Fecal transplants also pose some risks for safety and adoption may be a challenge.
What is precision microbiome editing?
The BIOME team is focused on developing both tools and real-world applications in precision microbiome editing. Precision microbiome editing is the use of genome-editing tools to edit the genes of microbes while they are in their natural environment, that is, within a microbiome.
“We’re building tools for being able to implement really precise control of these communities,” says Rubin. “We have a couple main goals for this toolset. One is to really understand communities better. As Brady mentioned, we have a bunch of guesses at genes important to microbiome function. But we have limited proof, because we don’t have the tools to test these guesses. The other key goal is to make changes to genomes to improve health, decrease emissions from livestock, or get other outcomes we want.”
Unlike an antibiotic, genome editing is highly precise, able to target not just a specific microbe in a community, but a specific part of a specific gene within that microbe. For example, if a species of bacteria in a gut microbiome is producing a compound that creates inflammation, a genome editing therapy could knock out the gene that encodes for that compound, or edit the gene to a known healthier state, while leaving the rest of the microbiome alone.
What is the BIOME team working on right now?
“Our early proof of concept work has shown that we can actually use the tools we’ve developed to perform targeted editing within a complex natural microbiome,” says Cress.
This will allow the team to understand the function of genes within that community by measuring the effect of a broken gene.
“We’ve also shown that we can use targeted editing to exert control within a microbiome. So for example, we can go into a microbiome and put a molecular tag on a specific kind of bacterium within that microbiome, and we can use that tag to increase the quantity of that bacterium relative to the rest of the community members,” says Cress. “And in fact, we’ve shown that we can do this on a species closely related to many other strains in that same community, highlighting the level of precision we achieve.”
Read more: